Molecular dynamics simulation of HSA-ligand complex for 50 ns time

Molecular dynamics simulation of HSA-ligand complex for 50 ns time

Unravelling Binding of Human Serum Albumin with Galantamine: Spectroscopic, Calorimetric, and Computational Approaches

Molecular Docking and Dynamics Simulation of Several Flavonoids Predict Cyanidin as an Effective Drug Candidate Against SARS-CoV-2 Spike protein - Abstract - Europe PMC

Molecular dynamics simulation of HSA–DAPG complex for 50 ns time scale

a) HSA-HA binding energy vs. time average for complex 1 (constant line
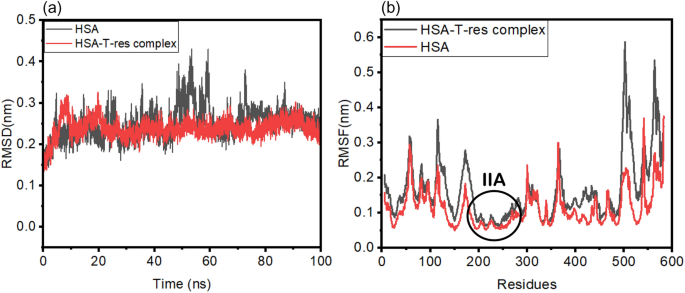
Investigating binding dynamics of trans resveratrol to HSA for an efficient displacement of aflatoxin B1 using spectroscopy and molecular simulation

Binding of Apo and Glycated Human Serum Albumins to an Albumin-Selective Aptamer-Bound Graphene Quantum Dot Complex

Ligand-protein complex poses after 20 ns molecular dynamics (MD)

Full article: Discovery of Potential Inhibitors for SFRP3: Ligand-Based 3D Pharmacophore, Virtual Screening, Molecular Docking, and Dynamics Studies
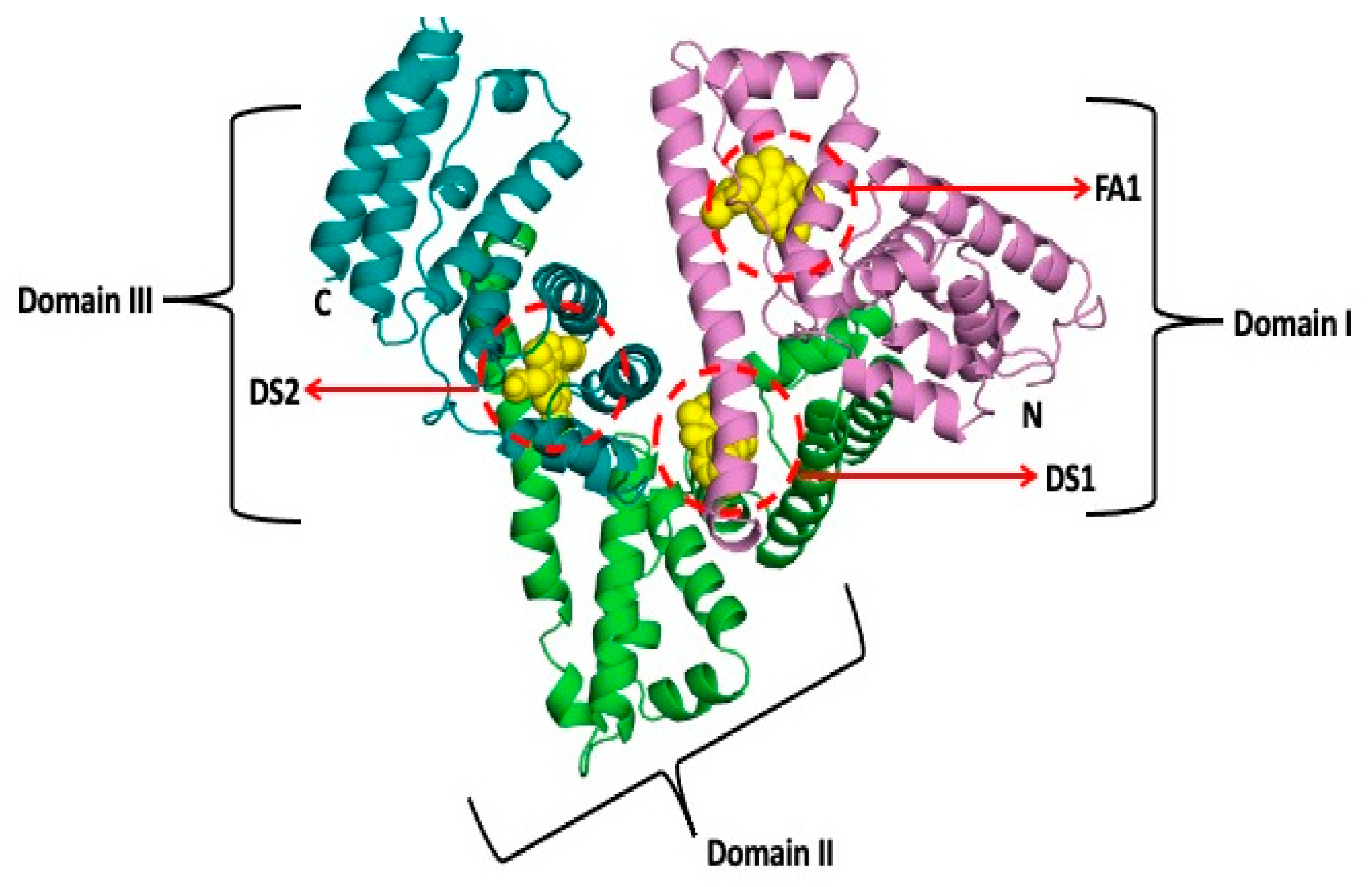
Molecules, Free Full-Text

IJMS, Free Full-Text


