Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes
Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes
Metabolism is a term that describes all the chemical reactions that are involved in keeping a living organism alive. Diseases related to metabolism – such as obesity, heart disease and diabetes – are a major health problem in the Western world. The causes of these diseases are complex and include both environmental factors, such as diet and exercise, and genetics. Indeed, many genetic variants that contribute to obesity have been uncovered in both humans and mice. However, it is only dimly understood how these genetic variants affect the underlying networks of interacting genes that cause metabolic disorders. Measuring gene activity or expression, and tracing how genetic instructions are carried from DNA into RNA and proteins, can reliably identify groups of genes that correlate with metabolic traits in specific organs. This strategy was successfully used in previous studies to reveal new information about abnormalities linked to obesity in specific tissues such as the liver and fat tissues. It was also shown that this approach might suggest new molecules that could be targeted to treat metabolic disorders. A brain region called the hypothalamus is key to the control of metabolism, including feeding behavior and obesity. Hasin-Brumshtein et al. set out to explore gene expression in the hypothalamus of 99 different strains of mice, in the hope that the data will help identify new connections between gene expression and metabolism. This approach showed that thousands of new and known genes are expressed in the mouse hypothalamus, some of which coded for proteins, and some of which did not. Hasin-Brumshtein et al. uncovered two genetic variants that controlled the expression of hundreds of other genes. Further analysis then revealed thousands of genetic variants that regulated the expression of, and type of RNA (so-called "spliceforms") produced from neighboring genes. Also, the expression of many individual genes showed significant similarities with about 150 metabolic measurements that had been evaluated previously in the mice. This new dataset is a unique resource that can be coupled with different approaches to test existing ideas and develop new ones about the role of particular genes or genetic mechanisms in obesity. Future studies will likely focus on new genes that show strong associations with attributes that are relevant to metabolic disorders, such as insulin levels, weight and fat mass..
In depth characterization of gene expression in the mouse hypothalamus will facilitate understanding of the molecular pathways that affect metabolic traits and discovers new genes associated with these pathways.

The regulatory landscape of multiple brain regions in outbred heterogeneous stock rats
Identification of putative regulatory regions and transcription factors associated with intramuscular fat content traits, BMC Genomics
Identification of putative regulatory regions and transcription factors associated with intramuscular fat content traits, BMC Genomics
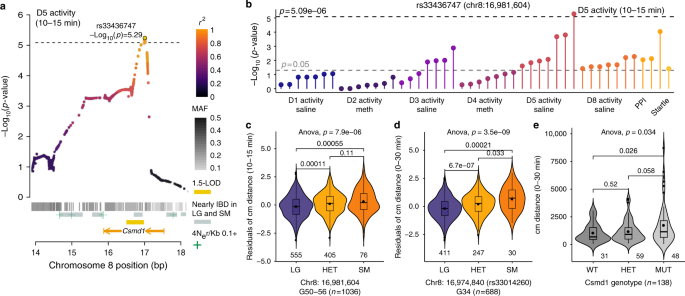
Genome wide association analysis in a mouse advanced intercross line

Simultaneous quantification of mRNA and protein in single cells reveals post-transcriptional effects of genetic variation

The regulatory landscape of multiple brain regions in outbred heterogeneous stock rats

Integration of feeding behavior by the liver circadian clock reveals network dependency of metabolic rhythms

The regulatory landscape of multiple brain regions in outbred heterogeneous stock rats

Where Are the Disease-Associated eQTLs? - Abstract - Europe PMC

PDF) Saturating the eQTL map in Drosophila melanogaster: genome-wide patterns of cis and trans regulation of transcriptional variation in outbred populations

mRNA Metabolism in Cardiac Development and Disease: Life After Transcription

Conserved multi-tissue transcriptomic adaptations to exercise training in humans and mice - ScienceDirect

Genome wide association analysis in a mouse advanced intercross line